Reading Molecular Data in JavaScript with PDB
The RCSB has a really great library of data for thousands of proteins in its Protein Data Bank, but sadly the formats that they offer are ancient. Both PDB and mmCIF formats use character indices to describe data fields, making them more suited to be read into FORTRAN than transferred over the modern web. After running into this problem enough times myself, I’m releasing the open source parse-pdb package in an attemp to save other people some time wrangling while loops and substring calls.
Getting Started
You can install the library just like you would any other npm module:
npm install --save parse-pdb
From there you’ll just import the module and call it with a string containing an entire PDB file. Node’s fs modules is nice for reading this info in from a file, but if you want to get it directly from RCSB or any other way it works too.
const parsePdb = require('parse-pdb');
const { readFileSync } = require('fs');
const pdbString = readFileSync('./3aid.pdb', 'utf8');
const parsed = parsePdb(pdbString);
// parsed now contains a nice javascript object with all the 3AID PDB data!
A Returned Format That’s Actually Useable
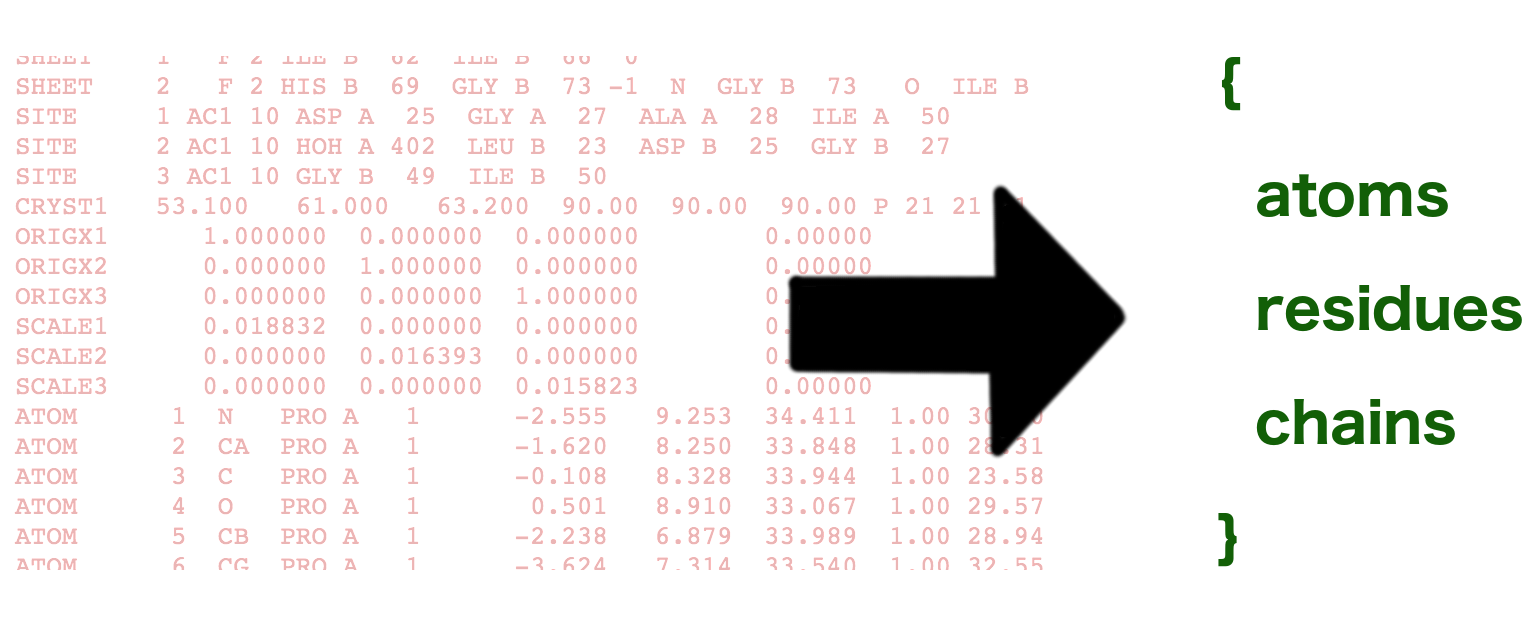
The object that you get back, called parsed in the example above, contains the basic data from the PDB more or less as-is, but it also has some more user-friendly derived data as well.
The basic data is parsed from the ATOM and SEQRES entries in the PDB. These are both arrays of objects that contain every data value in the row as a key/value pair. See the output format documentation for a list of every property and its type.
console.log(parsed.atoms); // [ { serial: 1, ... }, ...1845 others ]
console.log(parsed.seqRes); // [ { serNum: 1, ... }, ...1845 others ]
In addition, I’ve also parsed out residues and chains for convenience and given them references to their lower level elements. For example, you can get residues in a chain or atoms in a residue:
const residue = parsed.residues[0];
const residuesAtoms = residue.atoms; // array of 9 objects from parsed.atoms
const chain = parsed.chains.get(residue.chainID);
const siblingResidues = chain.residues; // array of 99 residues
Let’s Make Protein Data Easier to Work with on the Modern Web
I hope this parser can be of use to other people and help bridge the gap between scientific computing and the modern web. However, I’m more comfortable with the computing side than the science side, and this is just my guess at what others might find useful. If you’ve got any ideas for what might make this more useful to you, open an issue (or a pull request!) on the Github repository, or just drop a comment below.